Our Research
Bioinspired Chemistry and Materials
Our lab is developing and investigating biological systems that will enable the sustainable synthesis of valuable chemical compounds and fabrication of new types of functional materials through biological design.
Current Research Interests
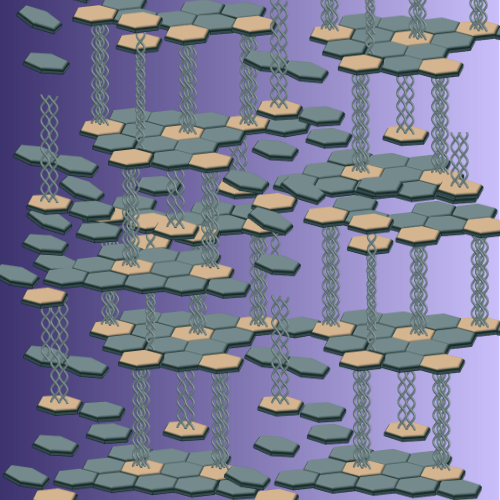
Biological Self-organization
We are interested in harnessing biological principles of self-organization for the design of genetically programmable materials for a range of applications.
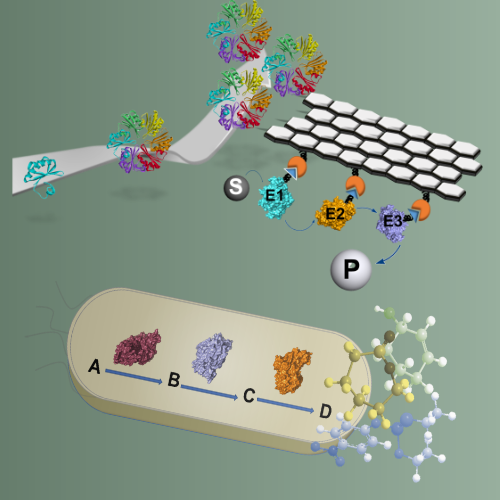
Biomanufacturing
We aim to explore and utilize the tremendous diversity of enzyme reactions performed by cells for the design of sustainable in vivo biosynthetic and in vitro biocatalytic routes to valuable chemical compounds
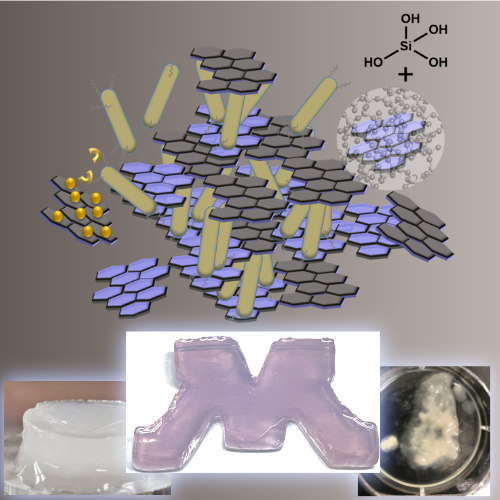
Biomineralization
Inspired by the extraordinary hierarchical composite materials made by living systems, we seek to adopt these mechanisms for the bottom-up design of new types of genetically programmable, functional biocomposites.
