Pumilio proteins: We study Pumilio proteins, a family of regulators that bind certain mRNAs with exquisite specificity and repress their expression. Pumilio proteins have diverse biological roles in development, stem cells, and fertility. Pumilio also controls neurological processes including motor neuron function, learning and memory formation. Our goals are to identify the mRNAs that Pumilio proteins regulate and determine the molecular mechanism of this regulation. To accomplish this, we use a combination of biochemistry, genetics, bioinformatics, transcriptomics, and high throughput assays in multiple organisms including humans and Drosophila. This research has direct impact on genetic mechanisms controlling development, neurological function, neurodegeneration, epilepsy, and cancer.
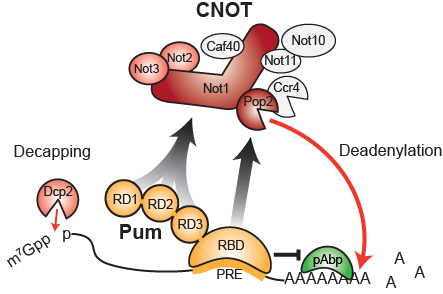
Model of Pumilio-Mediated Repression. The RNA-binding domain (RBD) of Pumilio binds to specific mRNAs that contain a Pumilio Response Element (PRE). Pumilio Repression Domains (RD1, 2, and 3) recruit the CCR-Not (CNOT) complex to accelerate deadenylation and decapping of the mRNA, and the RBD antagonizes poly-Adenosine binding protein, resulting in translational repression and mRNA destruction. Image derived from Arvola et al., Nucleic Acids Research, 2020.
Deadenylases: Ribonucleases play critical roles in regulating mRNAs. We focus on deadenylases, which are specialized ribonucleases that degrade the poly(Adenosine) tails of mRNAs. Regulation of poly(A) tail length is a critical control point for translation and mRNA degradation in a wide variety of biological contexts. Indeed, we found that specific deadenylases play a central role in Pumilio-mediated regulation. Pumilio proteins enhance deadenylation of the mRNAs they bind by directly recruiting the deadenylase enzyme complex.
The versatility of regulation by deadenylation is greatly expanded in higher eukaryotes through diversification of deadenylases. Humans possess twelve deadenylase orthologs. Genetic analysis indicates that each deadenylase controls unique biological functions including cell division and growth, metabolism, development, bone morphogenesis and anti-viral responses. We are exploring the questions: how many active deadenylases are there, do their catalytic activities differ, which mRNAs do they act upon, and how are their activities controlled? This research has broad relevance to fertility, development, and metabolism with relationships to obesity and cancer.
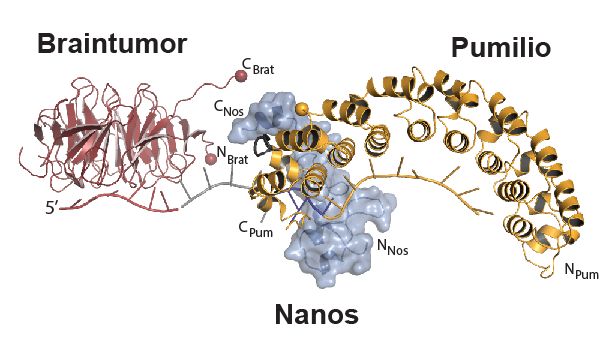
Pumilio collaborates with RNA-binding proteins Braintumor and Nanos to regulate the fate of certain mRNAs. Image derived from Arvola et al., RNA Biology, 2017.
Collaborations: Collaboration is a crucial hallmark of modern multi-disciplinary research. Our research capabilities and accomplishments have been greatly facilitated through collaborations with talented structural, computational, and developmental biologists. The University of Minnesota provides a rich environment for our research, which is promoted through the Minnesota RNA Supergroup.
We use CRISPR/Cas9 genome engineering to study the function of RNA-binding proteins in the fruit fly Drosophila melanogaster. Image courtesy of Robert Connacher.
Research Support. Our research is generously supported through grants from the following sources: