Candida albicans is the most frequently isolated fungal pathogen of humans, affecting immunocompromised patients ranging from premature infants to AIDS sufferers. Systemic infections have an attributed mortality of 30-50%. Although many properties have been shown to contribute to virulence in animal studies, its pathogenesis is not well understood. Analysis of the genome has been undertaken to provide researchers with more tools to investigate Candidiasis.
C. albicans is a diploid organism which has eight sets of chromosome pairs. Its genome size is about 16 Mb (haploid), about 30% greater than S. cerevisiae (baker’s yeast). In 1996, the Stanford Genome Technology Center undertook the sequencing of the C. albicans genome with the funding provided by the Burroughs Wellcome Fund and the National Institute for Craniofacial and Dental Research. Sequencing was completed to a level of 10.7x in 2004. Diploid and haploid assemblies are available at http://www.candidagenome.org/
The physical map of Candida albicans
Description of the physical map of Candida albicans
The physical map of Candida albicans consists of eight sets of overlapping fosmid clones which cover the eight chromosomes. The map is based on the macrorestriction map prepared by Chu, et al3 using the 8-base-pair specific restriction enzyme SfiI. The fosmid contigs are used to order and orient the haploid sequence contigs from the Candida genome project carried out by Jones, Scherer, Agabian, Davis, and their colleagues4. The map is being constructed as a joint venture between the University of Minnesota, the Chiba University Institute for Pathogenic Fungi and Microbial Toxicoses in Chiba, Japan, and the Biotechnology Research Institute of the National Research Council in Montreal, Canada (candida.bri.nrc.ca/candida/index.cfm).
The map is a Sequence-Tagged Site map. Probes which hybridize with more than one fosmid are used to generate fosmid contigs, and the contigs are assigned to chromosomes based on the hybridization of the probes to CHEF separations of chromosomes and SfiI digests. Sequenced probes are used to align the sequence contigs along the fosmid contigs. A sample of the map of chromosome 5 is shown for illustration.
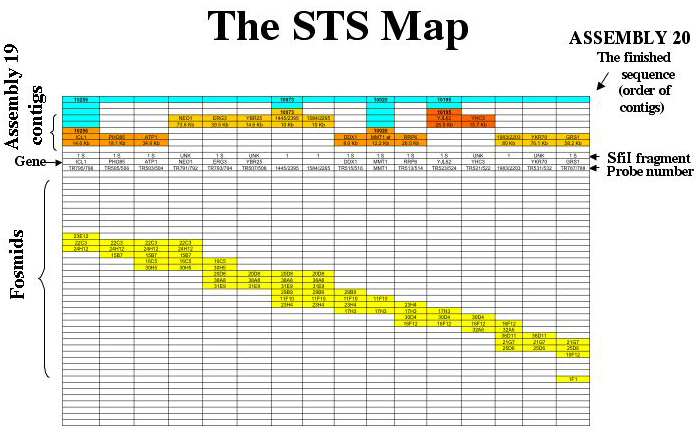
The map of each chromosome is arranged so that each column corresponds to a single data point. The identifiers for these data are as follows: On the map each column corresponds to a probe (site). The top line (blue) shows the supercontigs constructed at the Biotechnology Research Institute in Montreal. The sequence contigs are shown next in orange. If a known gene served as probe (or has been identified as being all or partly encoded by a probe) it is shown on the contig, and its position on the sequence contig (numbered as on the Assembly 19 server) is given below each gene name. The SfiI fragments (which also identify the chromosomes) are shown below the contigs. Probes which hybridize with more than one chromosome are listed as “MULTI”.
The gene names are listed next. Many probes have not been sequenced; the gene category is left blank for them. The centromeres as identified by Sanwal et al5 are listed in the “gene” row.
The probe is listed next. The probes are named according to an internal data base. When there is a break between fosmid contigs, the probe line contains a blue box labeled “break”.
The fosmid contigs are shown below. Each box labeled with a fosmid identifying number indicates that the probe for that column hybridizes with that fosmid. The fosmids coded red are those that are hit by a probe for the subtelomeric repeat. Thus, the ends of the chromosomes are marked by red fosmids. The fosmids coded violet are hit by non-telomeric probes that hit several chromosomes, for example, the Major Repeat Sequence (MRS) probes. The fosmids containing unique DNA (those that show hybridization only with probes that hybridize to a single chromosome on a CHEF gel) are coded yellow. In order to keep the page a reasonable size, the fosmid contigs are stepped.
The fosmid contigs shown here are the “tiling sets”, edited for clarity and usefulness. For the complete data set for a chromosome (including, for example, all the fosmids that hybridize with a particular probe, the fosmids that contain both unique DNA and a repetitive sequence, etc.) send an email to magee@umn.edu. The fosmids adjacent to specific sites of repetitive DNA (e.g., the MRS on any given chromosome) are available in a special database.
Sequenced-tagged sites are short DNA sequences which serve to locate larger clones
on a chromosome. For this map, colony hybridization is used to assign probes
to particular members of the fosmid library (3840 clones). The probes are
simultaneously hybridized to CHEF separations of chromosomes and SfiI fragments.
For a full description, see Chibana1.
MULTI
Probes which hybridize with several chromosomes are telomere-specific probes
(ex: 1972), probes for the MRS, probes which contain all or part of a retrovirus LTR,
or probes containing all or part of the conserved region of a multi-gene family.
REFERENCES
1. Chibana, H., B.B. Magee, S. Grindle, Y. Ran, S. Scherer, and P. T. Magee. 1998. A physical map of Chromosome 7 of Candida albicans. Genetics 149:1739-1752.
2. Chibana, H., N. Oka, H. Nakayama, T. Aoyama, B.B. Magee, P.T. Magee, and Y. Mikami. 2005. Sequence finishing and gene mapping for Candida albicans chromosome 7, and systenic analysis against Saccharomyces cerevisia genome. Genetics.
3. Chu, W.S., B.B. magee, and P.T. magee. 1993. Construction of an SfiI Macrorestriction map of the Candia albicans genome. Journal of Bacteriology 175:6637-51.
4. Jones, T., N.A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B.B. magee, G. newport, Y.R. Thorstenson, N. Agabian, P.T. Magee, R.W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A 101:7329-34.
5. Sanyal, K., M. Baum, and J. Carbon. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A 101:11374-9.